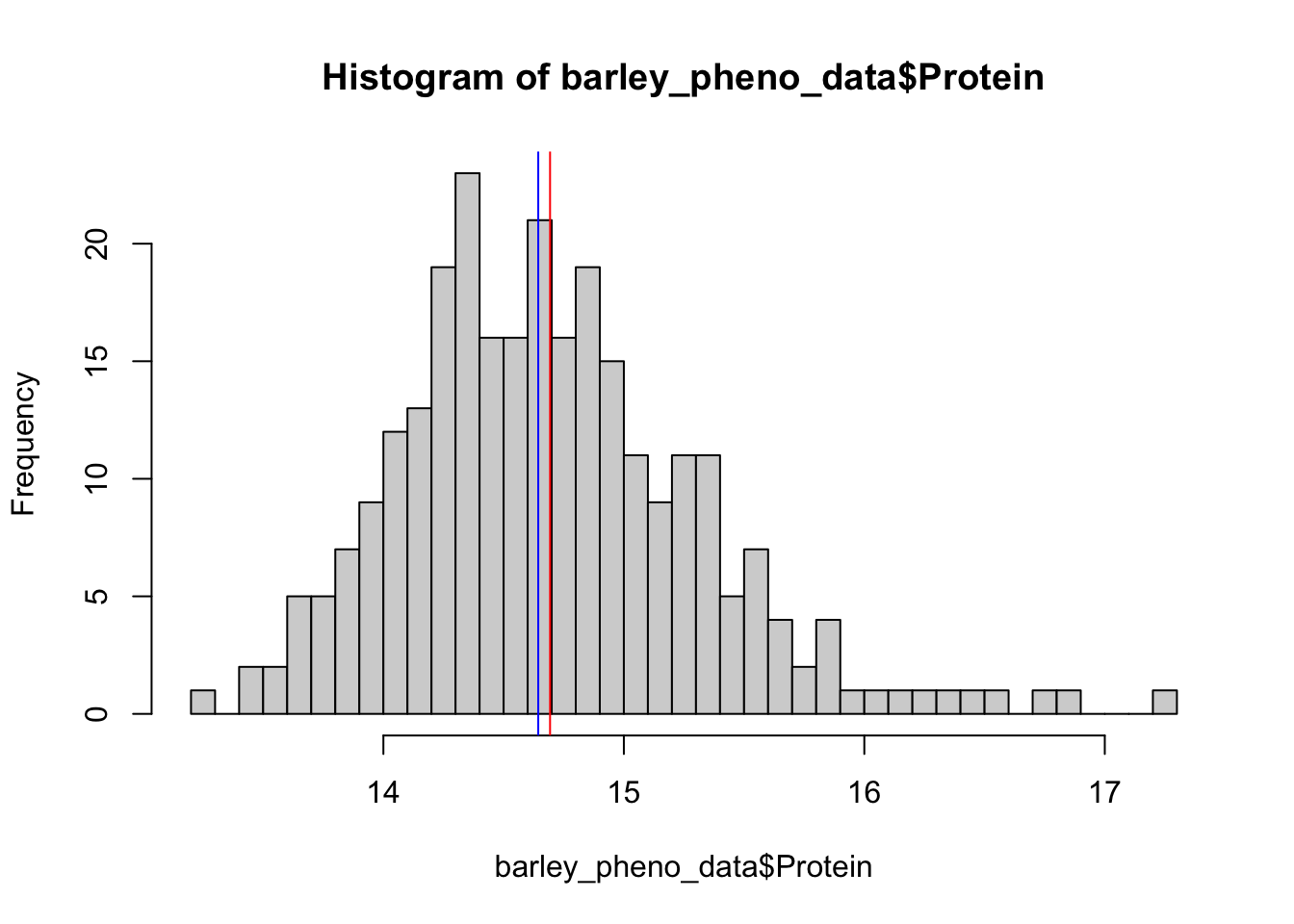
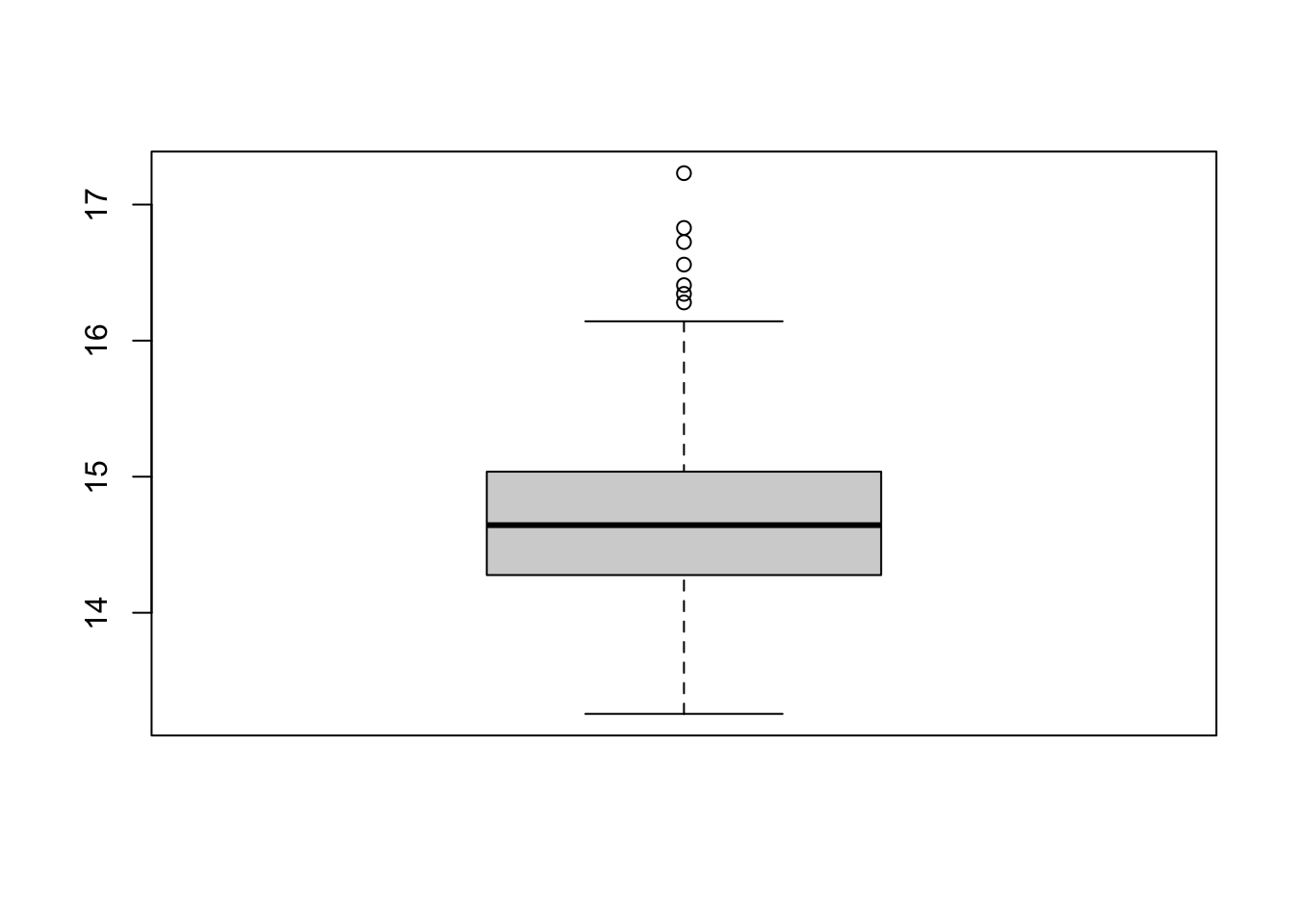
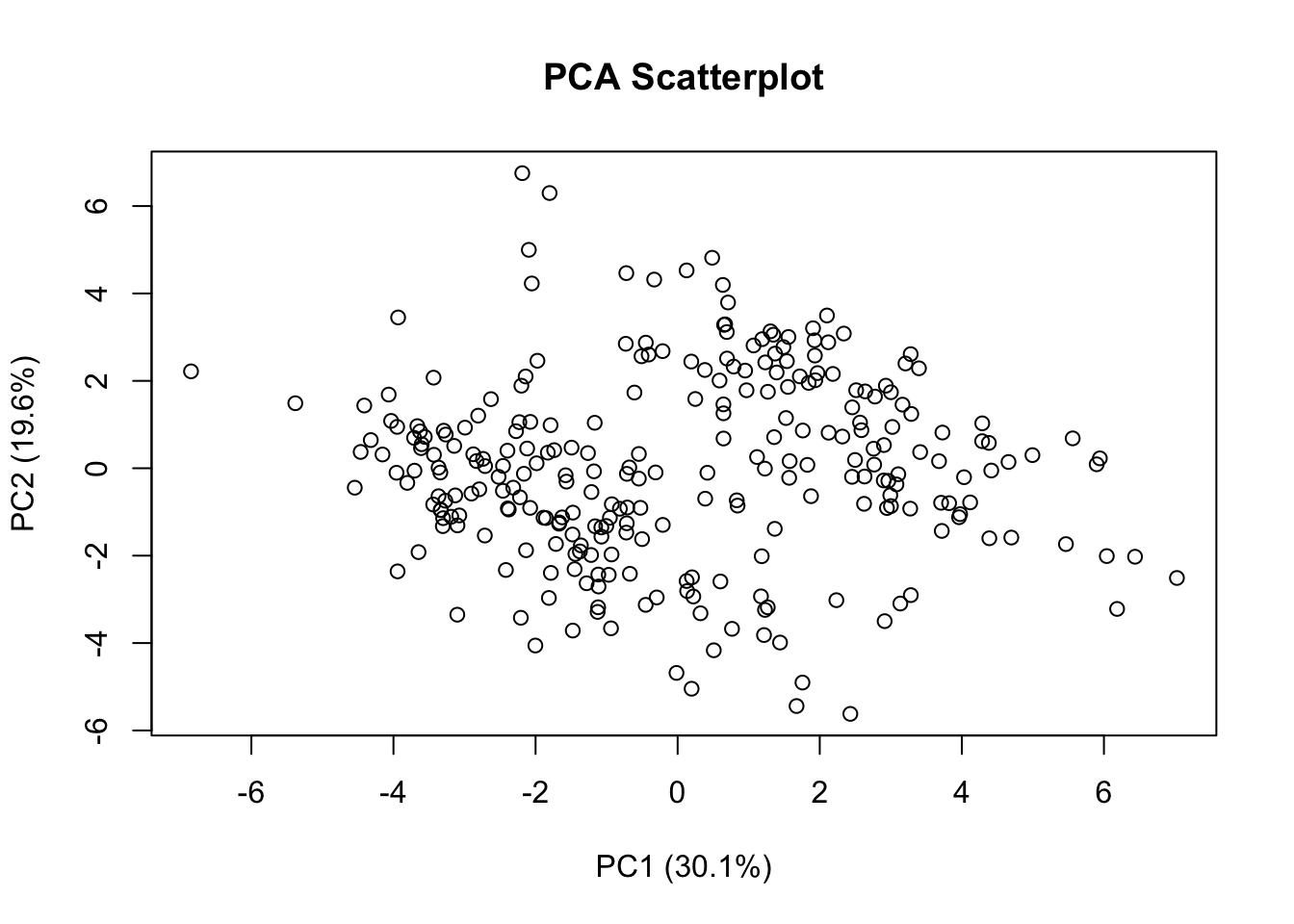
Rows: 275
Columns: 24
$ Taxa <chr> "G1", "G2", "G3", "G4", "G5", "G7", "G8", "G9", "G12", "G1…
$ Area <dbl> 22.72177, 23.07877, 19.93612, 23.32083, 19.54859, 22.96829…
$ B_glucan <dbl> 6.848584, 7.430943, 4.012621, 6.091926, 7.307811, 7.267738…
$ Circularity <dbl> 1.887915, 1.780070, 1.555492, 2.300471, 1.896487, 1.752493…
$ Diameter <dbl> 5.366522, 5.403555, 5.034355, 5.420909, 4.981714, 5.422541…
$ DTH <dbl> 75.89584, 70.17532, 74.19461, 74.39462, 77.66037, 72.65420…
$ Fe <dbl> 29.67685, 31.59088, 34.15825, 31.44544, 30.31127, 30.61605…
$ FLA <dbl> 16.586760, 7.966124, 7.592350, 18.753322, 16.532845, 18.40…
$ FLH <dbl> 82.77116, 62.36145, 61.81653, 69.77661, 64.38496, 78.08801…
$ GpS <dbl> 44.70886, 25.25222, 25.72167, 66.23116, 54.63221, 26.63211…
$ GWS <dbl> 1.6156277, 1.0563208, 0.9516462, 2.5202010, 1.2044795, 1.0…
$ GY <dbl> 1.3747605, 1.3735596, 0.9054266, 0.7614383, 0.8827177, 2.0…
$ HW <dbl> 57.65487, 64.41055, 69.41048, 55.35590, 53.65940, 64.98411…
$ Length <dbl> 10.542981, 10.106793, 8.565790, 11.920853, 9.781558, 9.968…
$ Length_Wid <dbl> 3.313525, 3.013035, 2.524445, 3.833381, 3.305421, 2.978634…
$ PdH <dbl> 91.09761, 63.65600, 68.75482, 69.50663, 64.10002, 80.59519…
$ PdL <dbl> 8.1320940, 1.0341930, 6.5555050, 0.9639243, -0.6416490, 2.…
$ Perimeter <dbl> 29.07010, 28.33654, 24.57919, 32.21194, 27.02846, 28.03643…
$ PH <dbl> 96.76140, 70.56720, 77.16486, 79.22446, 71.78375, 88.70248…
$ Protein <dbl> 14.06372, 14.16090, 15.23294, 14.34877, 14.46224, 14.31356…
$ SL <dbl> 5.343668, 6.804813, 8.074976, 10.039594, 7.526454, 8.31457…
$ TKW <dbl> 28.48964, 35.00164, 31.95561, 27.27054, 24.64983, 33.83401…
$ width <dbl> 3.251696, 3.439623, 3.455571, 3.204069, 3.000305, 3.429043…
$ Zn <dbl> 31.21179, 34.21217, 25.50724, 32.24918, 33.58773, 33.92710…